Experimentally determined binding constants represent a cornerstone of thermodynamic characterization for supramolecular chemistry. This workshop aims to update participants on the cutting-edge tools and techniques for ascertaining high quality binding constants from spectroscopic titrations (UVvis, NMR, etc.).
Open to all undergraduates, graduate students, post-docs, and professors, the workshop will occur in two 1.5-hour sessions on Sunday, December 15, 2024, starting at 2:00 pm. The cost is $50. A limited number of scholarships are available for those that otherwise would not be able to participate.
You will have the option to register for this workshop during the normal conference registration, but the registration fee itself will be arranged separately. The fee for the workshop is $50. If you have selected that you will be attending the workshop, you will receive an email to arrange for the registration fee from the workshop leaders (Amar Flood and Douglas Vander Griend).
The learning objectives are:
- Understand the quantitative aspects of chemical equilibria in supramolecular and biochemical systems.
- Build and comprehensively analyze spectroscopic titration data.
- Use appropriate methodological tools to model data.
- Design optimal experiments for binding constant quantification.
- Report results in communally beneficial ways.
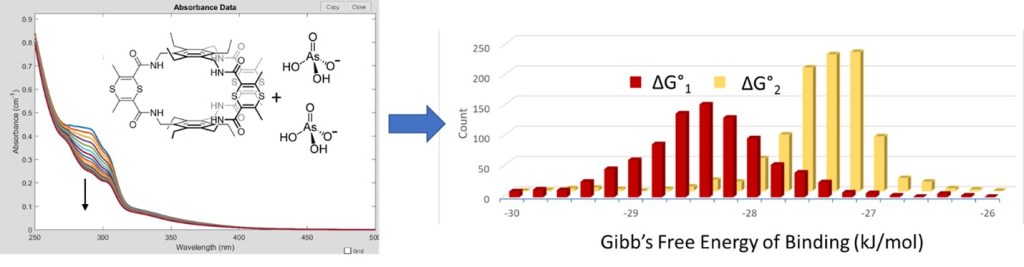
Key questions around which this workshop is built:
- What important global questions rely on Supramolecular Chemistry?
- Why do supramolecular chemists care about binding constants?
- How has data science evolved to help model, quantify, and catalog binding constants?
- What kind of experimental data can help quantify binding constants?
- What are some key strategies for getting high quality titration data?
- Which solution compositions best help quantify the binding constant(s)?
- Why does modeling measured data provide the best way to quantify binding constants?
- What computer tools exist to help model spectroscopic titration data?
- How is the uncertainty of the binding constants best understood and quantified?
- How many distinct chemical species occur throughout my titration?
- How many distinct chemical species are needed to model my titration dataset?
- How can I tell if one model is better than another?
- How can we help each other to follow best practices when quantifying binding constants?